Documentation
This documentation provides detailed information on how we collected data and constructed molecular pathways, as well as how the data can be used for further analysis.
What are rare CNVs
Copy number variants (CNVs) are structural genomic alterations involving DNA segments typically larger than 1 kilobase (kb). They contribute to human genetic diversity and have been implicated in various neurodevelopmental and neuropsychiatric disorders, including intellectual disability (ID), autism spectrum disorder (ASD), bipolar disorder (BD), and schizophrenia (SCZ). (Feuk et al., 2006; Redon et al., 2006; Rees et al., 2021)
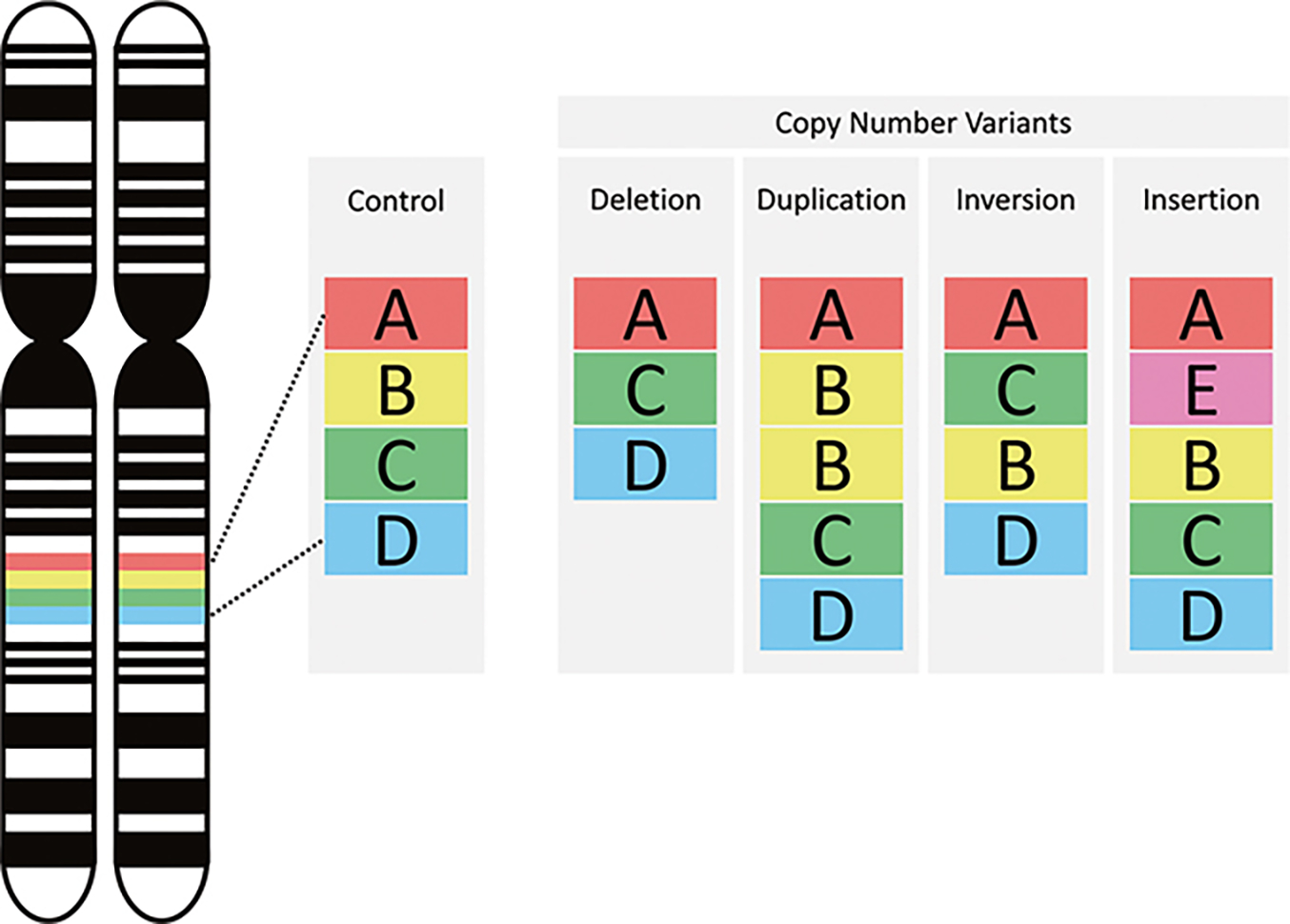
CNV selection criteria
We selected CNVs based on the availability of curated molecular pathways in the WikiPathways Rare Diseases community. In total, this resource includes 35 unique molecular pathways.
Most CNVs represented here are recurrent (i.e., involve the same genomic breakpoints) and are known to have a high prevalence among individuals with psychiatric disorders such as schizophrenia, as reported by Marshall et al., 2017. However, this resource is not limited to schizophrenia associated CNVs.
Data linked to each CNV
Diseases associated with deletions or duplications in specific genomic regions were collected from established databases such as OrphaData and OMIM.
Each CNV was linked to these resources using their unique identifiers ORPHAcodes and OMIM IDs, respectively.
When information was missing from these databases, we conducted literature research to supplement the data.
Disease associated information
From OrphaData we retrieved:
- Disease descriptions
- Prevalence
- Associated OMIM IDs
- Phenotypic features labeled as Very frequent (99–80%)
These phenotypic features were mapped to the Human Phenotype Ontology (HPO), providing structured and computable clinical data.
Genes associated information
For gene level information, we queried the HGNC database to extract:
- Approved gene symbols
- Gene names
- Cross-references to external databases such as NCBI, Ensembl, and UniProt
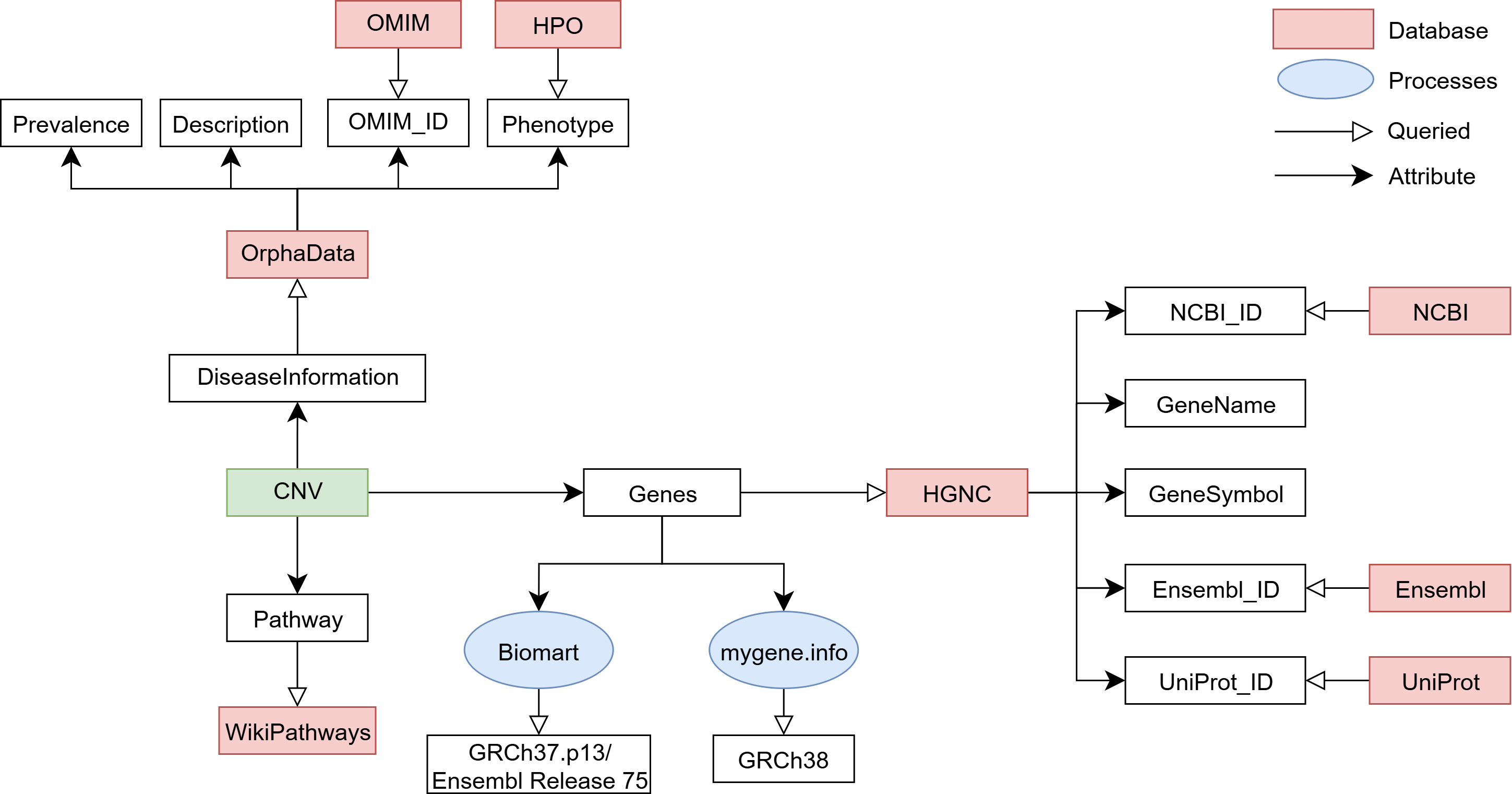
Molecular pathway construction
To build molecular pathways we used PathVisio together with BridgeDb and WikiPathways plugins.
If you are new to PathVisio, start by following this PathVisio setup tutorial to install the software and required plugins.
Once set up, you can learn how to create your own pathway using the WikiPathways Academy.
After creating your pathway, you can upload it to WikiPathways to share your knowledge with the community and even become a curator.
Further analysis with Cytoscape
You can download the entire copy number variants table from the main page.
| cnv | locus | chromosome | start | end | description | pubmed_id | genes_hgnc_symbol | genes_hgnc_name | genes_hgnc_id | genes_entrez_id | genes_ensembl_id | genes_uniprot_id | wikipathways_id | orphadata_orphacode | orphadata_cause | orphadata_definition | orphadata_prevalence | orphadata_phenotypes | orphadata_hpo_id | orphadata_omim_id |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1p36.33-p36.32 | 1p36.33-p36.32 | 1 | 0 | 2500000 | 1p36.33-p36.32 deletion or duplication… | - | OR4F5 | olfactory receptor… | HGNC:14825 | 79501 | ENSG00000186092 | Q8NH21 | WP5345 | 1606 | deletion | A rare chromosomal anomaly… | 15.0 (1–5 / 10 000)… | Pointed chin | HP:0000307 | 616975;607872 |
- Open Cytoscape or download it from cytoscape.org
- Go to File → Import → Network from File
- Select your CNV table (e.g. all_cnvs_table.xlsx or a filtered version)
- In the import dialog, set the appropriate columns:
- one column as Source Node (e.g.
cnv) - another as Target Node (e.g.
genes_hgnc_symbol)
- one column as Source Node (e.g.
- Click OK to import and view the network
- Extend the network using the cytargetlinker app with drug-, pathway-, or disease-related linksets
Licence
This content is licensed under the Commons Attribution 4.0 International (CC BY 4.0) licence. This means you are free to reuse the content in any way, including copying, distributing, displaying, or using it for commercial purposes, in any country or jurisdiction. The only requirement is that you give appropriate credit to us and to the original data sources we used (see citation guidelines below).
How to cite
How to cite our work
CNV website
CNV-Booklet: A comprehensive resource of Rare Copy Number Variants. Available on https://alexandra-valeanu.github.io/cnv-booklet/ (accessed on date).
Data version
You can cite the latest data version, or the specific version you used, from Zenodo:
Example citation for Release v_2025-07-22 (Harvard style): Alexandra Valeanu (2025) ‘alexandra-valeanu/cnv-data: Release v_2025-07-22’. Zenodo. doi: 10.5281/zenodo.16319401.
How to cite external data sources
Orphanet/Orphadata
Orphanet: an online rare disease and orphan drug data base. © INSERM 1999. Available on http://www.orpha.net. Accessed (accessed on date).
Orphadata: Free access data from Orphanet. © INSERM 1999. Available on https://www.orphadata.com. Data version (XML data version).
For further use of Orphadata, please consult their legal notice.
HGNC
Genenames.org: the HGNC resources in 2023 Ruth L Seal, Bryony Braschi, Kristian Gray, Tamsin E M Jones, Susan Tweedie, Liora Haim-Vilmovsky, Elspeth A Bruford Nucleic Acids Research (Database issue) NAR | EuropePMC | PubMed
Disclaimer
We are not affiliated with Orphanet or HGNC. We did not modify the data from Orphanet and HGNC.
The content in this booklet is provided for informational and research purposes only and is not intended as medical, legal, or professional advice. The information presented here is offered for improving the understanding of rare copy number variation syndromes.
Every effort has been made to ensure the accuracy and usefulness of the information contained in this booklet. However, the authors do not claim any liability arising from the use or misuse of this material. Use of the content is at the user’s own discretion and risk.